J Clin Aesthet Dermatol. 2023;16(2):44–49.
J Clin Aesthet Dermatol. 2023;16(2):44–49.
by Olga Marushchak, DO; Rebecca Yakubov, BHSc; Rose Yakubov, BHSc; and Gary Goldenberg, MD
Dr. Marushchak is with the Department of Internal Medicine at Mount Sinai Morningside-West in New York, New York. Dr. Goldenberg is with the Department of Dermatology at Icahn School of Medicine at Mount Sinai in New York, New York. Ms. Rebeeca Yakubov and Ms. Rose Yakubov are with McMaster University in Hamilton, Ontario.
ABSTRACT: Analysis of morphological characteristics for the diagnosis of melanoma remains a challenge. New technologies for the diagnosis and prognosis of melanocytic lesions have been emerging to ensure earlier and more accurate detection. In this article, we review multiple technologies that improve melanoma diagnostic accuracy such as electrical impedance spectroscopy, pigmented lesion assay, reflectance confocal microscopy, and gene expression profile tests. Keywords: Melanoma, diagnosis, electrical impedance spectroscopy, pigmented lesion assay, reflectance confocal microscopy, gene expression profile.
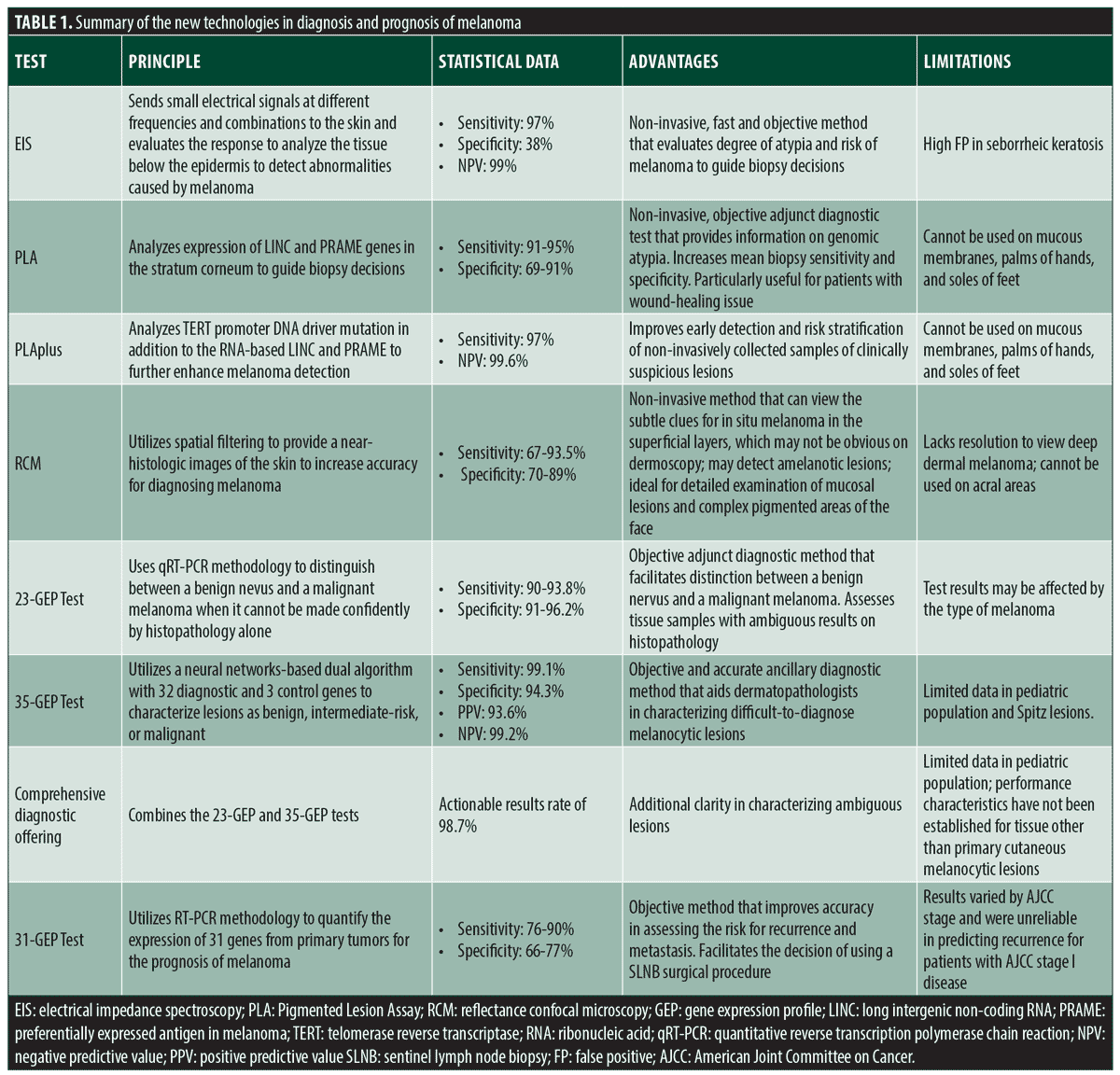
The rates of melanoma have been rising rapidly over the past few decades, the American Cancer Society estimates about 106,110 new melanomas will be diagnosed and about 7,180 people were expected to die of melanoma by the end of 2021.1 The emergence of new technologies for the diagnosis and prognosis of melanocytic lesions have helped reduce the proportion of individuals affected by melanoma. Electrical impedance spectroscopy (EIS) is a non-invasive Food and Drug Administration (FDA) approved technology, that is used to aid biopsy decisions in clinically challenging lesions.2 Similarly, pigmented lesion assay (PLA), a non-invasive technology, analyzes gene expression to guide biopsy decisions.3 Reflectance confocal microscopy (RCM), a non-invasive tool, provides near-histologic resolution images to facilitate the diagnosis of melanoma.4 Gene expression profile (GEP) tests use reverse transcription polymerase chain reaction (RT-PCR) methodology and quantify the expression of genes to distinguish melanoma from nevi and provide more accurate prognosis.5,6 This review summarizes the current technologies highlighting the principle, utility, advantages, and limitations for each technology (Table 1).
Electrical Impedance Spectroscopy
Nevisense (SciBase, Stockholm, Sweden) is a non-invasive tool approved by the United States (US) Food and Drug Administration (FDA) used to detect melanoma/atypical nevi to facilitate biopsy decisions. The device uses EIS to send small electrical signals at different frequencies and signal combinations to the skin and evaluates the response. The use of EIS allows for the detection of abnormalities caused by melanoma from the tissue below the epidermis. An AI classifier determines the degree of atypia and risk of melanoma by calculating a risk score between 0 to 10.7 Several studies have demonstrated that the addition of this device improved clinical accuracy of diagnosing melanomas. The SciBase International Melanoma Pivotal Study (SIMPS) showed that the EIS device detects malignant melanoma accurately and safely.8 The study was conducted internationally at 22 trial sites (17 clinics in Europe, 5 clinics in the US) and investigated 2,416 lesions on 1951 patients. The results demonstrated 97 percent sensitivity for 265 malignant melanomas: 94 percent sensitivity for 112 melanomas in situ, 99 percent sensitivity for 113 T1 melanomas, and 100 percent sensitivity for 40 T2-T4 melanomas. The results showed 38-percent specificity and a negative predictive value of 99 percent.8 Svoboda et al9 compared the diagnostic accuracy and biopsy decisions of clinicians with morphologic assessment alone and with an EIS score. An online survey presenting 45 pigmented lesions (28 benign, 17 melanoma) was completed by 164 dermatology trainees. The trainees were tasked with observing each image with a morphologic assessment alone and then with the corresponding EIS score (along with positive and negative predictive values) to determine if they would recommend biopsy. Overall, the results altered the individual biopsy decision in 24.3 percent of cases and the addition of the EIS score increased the mean sensitivity from 80.7 percent to 95.2 percent (P<0.001) and the mean specificity from 50.4 percent to 58.6 percent (P<0.001).9 Another study conducted by Rocha et al10 evaluated the effect of EIS measurement used in conjunction with sequential digital dermoscopy imaging (SDDI). SDDI is commonly used to evaluate suspicious melanocytic lesions. However, SDDI uses an assessment of successive dermoscopic images to detect melanoma, delaying the diagnosis by three months and requiring patients to return to the clinic. The study used a combined protocol, where all lesions underwent SDDI and EIS at baseline and the follow-up schedule was based on the EIS score. The score of 0 to 3 indicated benign lesion and did not require follow-up with SDDI; the score of 4 to 6 indicated benign lesion but required SDDI and possibly excision if changes are detected at the follow-up visit; the score of seven or more indicated possible melanoma and require excision without additional SDDI. The results demonstrated the combined protocol to have 100-percent sensitivity (95% confidence interval (CI) 54.1-100) and reduce the need for routine SDDI by 46.9 percent (95% CI 39.0-54.9).10
Although the EIS device demonstrated high sensitivity, specificity, and accuracy in the distinction of benign lesions of the skin from melanoma, it should be noted that the test inaccurately classified high proportion of seborrheic keratoses as positive. However, dermatologists or clinicians trained in the diagnosis of skin cancer should be able to recognize seborrheic keratosis clinically and avoid applying EIS to these lesions.8
The Pigmented Lesion Assay
PLA is a non-invasive gene-expression test which allows clinicians to rule out melanoma. PLA works by tape-stripping lesions to obtain the stratum corneum from which RNA is isolated. Once RNA is isolated, expression levels of long intergenic non-coding RNA 518 (LINC) and preferentially expressed antigen in melanoma (PRAME) genes are assessed. A PLA test result is positive if LINC, PRAME or both target genes are detected, as these molecular findings were found to correspond with histopathology findings of melanoma in situ or invasive primary melanoma in seven percent, 50 percent, and 93 percent, respectively.11 A positive PLA result would indicate a biopsy and histopathology, a negative result would indicate a clinical follow-up and no biopsy.12
A study conducted by Gerami et al13 validated that expression of LINC and PRAME accurately classify pigmented lesions for cutaneous melanoma. Pigmented lesions (157 training and 398 validation samples) were evaluated for LINC and PRAME gene expression using adhesive patch biopsy. Results were compared with histopathological diagnoses established by three experienced dermatopathologists. In 398 validation samples (87 melanomas and 311 non melanomas), PLA accurately differentiated melanoma from nonmelanoma cases with a sensitivity of 91 percent (95% CI 83%-96%) and a specificity of 69 percent (95% CI 64%-74%)
Ferris et al14 studied the real-world clinical performance of PLA and the influence PLA has on physician diagnosis. The study assessed 381 patients, with 51 PLA(+) cases and 330 PLA(-) cases. Of the 51 PLA(+) results, 19 (37%) were confirmed to be melanomas. Of the 330 PLA(-) test results, three lesions were subjected to follow-up biopsies, and histopathologically diagnosed as nonmelanomas. As a result, these data points demonstrate an overall estimated 95 percent sensitivity and 91 percent specificity of the PLA.
A large US registry study conducted by Brouha et al13 examined how PLA can help clinicians with determining the genomic risk factor of a lesion and guide clinicians with biopsy decisions. Overall, out of 3418 clinically suspicious melanoma skin lesions assessed by PLA, 324 lesions (9.48%) were PLA(+), and 3094 lesions (90.52%) were negative. Out of all PLA(+) lesions, 97.53 percent were surgically biopsied, while 99.94 percent of PLA(-) cases were clinically monitored and not biopsied.
A study by Ferris et al15 that provided a 12-month follow-up of PLA(-) cases demonstrated that from the 738 pigmented PLA(-) lesions that were followed up, only 1.8 percent of the lesions were biopsied (none of which had a histopathologic diagnosis of melanoma) within the 12 month follow-up period.
Ferris et al also aimed to determine the utility of the PLA in biopsy decisions in another study. In the study, 45 (29 male and 16 female) board-certified dermatologists each evaluated 60 images of pigmented lesions and determined whether the lesion should be biopsied (first without PLA and then with the addition of PLA). Overall, mean biopsy sensitivity was increased from 95.0 percent to 98.6 percent (p=0.01), and the specificity increased from 32.1 percent to 56.9 percent (p<0.001) when used in combination with the PLA data.16
Due to its non-invasive nature, PLA may be particularly useful for patients with wound-healing issues, coagulopathies, tendency to develop hypertrophic scars, or lesions in cosmetically sensitive areas. However, the test has its limitations as it cannot be used on mucous membranes, palms of hands, and soles of feet.17
A more advanced assay, PLAplus (DermTech, La Jolla, California) combines gene expression with gene mutation analyses to further enhance melanoma detection. The new test introduced telomerase reverse transcriptase (TERT) promoter DNA driver mutation analysis in addition to the RNA-based LINC and PRAME.18 TERT mutations are commonly found in early-stage melanoma and have been detected in 73 percent of histopathologically confirmed melanoma. Jackson et al19 demonstrated that gene expression and mutation data improves ability to stratify risk non-invasively. One hundred and three clinically suspicious skin lesions for melanoma were evaluated using adhesive patches. All lesions were then surgically biopsied to compare genomic analyses with histopathologic diagnoses by three dermatopathologists. TERT mutations were detected in 70 percent of PLA-positive lesions histopathologically diagnosed as melanoma and in 4 percent of severely dysplastic nevi and non-melanoma lesions excluding severe histologic atypia. The addition of TERT mutation analyses demonstrated an increase of the PLA test’s sensitivity for melanoma from 93 to 97 percent, in addition to increasing its negative predictive value from 99.3 to 99.6 percent.
Reflectance Confocal Microscopy
In-vivo RCM is a non-invasive technology that examines skin with cellular resolution. Confocal microscopy uses spatial filtering to generate a horizontal view of the skin, displaying cellular and subcellular structures up to the level of the upper dermis onto a screen.20 Several studies have demonstrated that confocal microscopy can serve as an important adjunct in clinical exams, dermoscopy, and histopathology assessment, increasing the accuracy for diagnosing melanoma. Lan et al21 investigated the accuracy of RCM for diagnosing melanoma in a systematic review. The study included seven studies with a total of 1,111 lesions. Results demonstrated that the sensitivity of RCM for the diagnosis of melanoma was 67 percent (95% CI 0.51–0.81) and the specificity was 89 percent (95% CI 0.86–0.92). Xiong et al22 compared the accuracy of dermoscopy versus RCM for the diagnosis of malignant skin tumors. Eight published studies, involving 1,141 skin lesions, were analyzed at the per-lesion level using RCM and dermoscopy. The pooled sensitivity of dermoscopy for melanoma detection was 88.4 percent (95% CI 0.84-0.92), whereas the pooled sensitivity of RCM was 93.5 percent (95% CI 0.90-0.96). The pooled specificity of dermoscopy for melanoma detection was 49.1 percent (95% CI 0.84-0.92), whereas pooled specificity of RCM was 78.8 percent (95% CI 0.75-0.82). Evidently, RCM could be used to improve the detection of malignant melanoma due to the significantly greater diagnostic specificity compared to dermoscopy.22 Another systematic literature review conducted by Pezzini et al23 assessed the diagnostic accuracy of RCM for malignant melanoma according to study design, lesion type and diagnostic modality and included a comparison with dermoscopy. A total of 32 studies (7,352 lesions) were included in the meta-analysis. Results demonstrated a pooled sensitivity of 92 percent (95% CI 0.91-0.93) and specificity of 70 percent (95% CI 0.69-0.71). Additionally, the diagnostic accuracy of RCM was greater than that of dermoscopy as the specificity of RCM was 56 percent (95% CI 0.52-0.60) and dermoscopy was 38 percent (95% CI 0.34-0.42).
RMC should be used as an additional tool in combination with dermoscopy, as the two modalities have specific features that help them complement each other. RCM can view the subtle clues for in-situ melanoma in the superficial layers, which may not be obvious on dermoscopy. However, RCM lacks resolution to view deep dermal melanoma, which could be assessed with dermoscopy. Additionally, RCM uses refractive properties of subsurface structures and may detect amelanotic lesions, which may not be visible on dermoscopy. Lastly, RCM cannot be used on acral lesions due to thick keratinized stratum corneum, but it is ideal for detailed examination of mucosal lesions and complex pigmented areas of the face.20
Gene Expression Profile Tests
MyPath Melanoma (Castle Biosciences, Inc., Friendswood, Texas) is a 23-GEP test that uses quantitative reverse transcription polymerase chain reaction (qRT-PCR) methodology to distinguish between a benign nevus and a malignant melanoma when the distinction cannot be made confidently by histopathology alone.5 The expression of 23 genes (14 melanoma signature genes and 9 reference genes) is measured and the ratio of signature to reference gene RNA is assessed. The result is a numerical value ranging from -16.7 to 11.1. Scores ranging from -16.7 to -2.1 are classified as likely benign, -2.0 to -0.1 are classified as indeterminate, 0.0 to +11.1 are classified as likely malignant. Several studies have validated that the 23-GEP test reliably differentiates uncertain melanocytic neoplasms. Clarke et al. conducted a prospective cohort study to assess the ability of the 23-GEP test to distinguish melanoma from benign nevi in clinically relevant lesions. A set of 1400 melanocytic lesions were diagnosed concordantly as benign or malignant by three experienced dermatopathologists. Each lesion was given a gene expression signature score. Results demonstrated that the test differentiated benign nevi from malignant melanoma with a sensitivity of 91.5 percent (95% CI 86.4%–95.2%) and a specificity of 92.5% (95% CI 90.0%–94.5%).24 These results are consistent with a retrospective cohort study of 437 malignant melanomas and benign nevi, which demonstrated that this test differentiated benign nevi from malignant melanoma with a sensitivity of 90 percent (95% CI 85–93%) and a specificity of 91 percent (95% CI 85–93%).5 In addition, multiple studies have compared the 23-GEP test results with actual patient outcomes, demonstrating a strong correlation. Ko et al25 assessed the performance of the test in detecting lesions with unequivocal signs of malignancy in a study of 182 samples (99 primary cutaneous melanomas with distant metastases and 83 melanocytic nevi). Results from the test differentiated melanoma from nevi with a sensitivity of 93.8 percent and a specificity of 96.2 percent.25 Next, Ko et al. compared the 23-GEP test results to histopathological diagnoses, development of local recurrence, sentinel lymph node metastases, and distant metastases. Archival formalin-fixed paraffin-embedded tissue sections of melanocytic lesions were examined. Out of 127 melanocytic lesions, 65 lesions were diagnosed as melanoma, while 62 lesions were diagnosed as benign by histopathology. The 23-GEP test results were malignant in 61 of 65 (93.8%) lesions and benign in 48 of 62 (77.4%). All 14 lesions that developed metastases were correctly identified by the test. This study demonstrated a strong correlation between the 23-GEP test results and clinical outcomes.26 The test results may be affected by the type of melanoma. In the subtypes of melanoma in situ where lesional melanocytes are distributed as single cells or widely scattered nests, the inclusion of excessive benign tissue could dilute tumor markers and potentially lead to false-negative results.24
DiffDx-Melanoma (Castle Biosciences, Inc., Friendswood, Texas) is a new 35-GEP test that utilizes a neural networks-based dual algorithm with 32 diagnostic and three control genes to characterize lesions as benign, intermediate-risk, or malignant.27,28 This test aids dermatopathologists in characterizing difficult-to-diagnose melanocytic lesions by providing a highly accurate, objective result.27 A study by Estrada et al27 validated the ability for the 35-GEP test to provide diagnostic clarity. The validation cohort consisted of 273 benign nevi and 230 melanomas. The 35-GEP test performed with high accuracy: 96.2 percent (95% CI 91.5-97.1%) sensitivity, 94.3 percent (95% CI 91.5-97.1%) specificity, 93.6 percent (95% CI 90.5-96.7%) PPV, and 99.2 percent (95% CI 98.1-100%) NPV. In all ages population, the test provided a clinically actionable result in over 96 percent of cases with an intermediate-risk result in 3.6 percent of cases. A study by Farberg et al. evaluated the clinical utility of the 35-GEP. Dermatopathologists (n=6) and dermatologists (n=14) were presented with 60 difficult-to-diagnose melanocytic neoplasms and asked to review each lesion pre- and post-35-GEP result. Results demonstrated that overall diagnostic confidence was increased in 51 percent of cases. In benign 35-GEP results, additional work-up requests were decreased (72.1%), while in malignant 35-GEP results, additional work-up requests were increased (45.6%).29 Due to limited data, potential limitations of the test may include lower accuracy in diagnosing Spitz lesions and lesions in pediatric population.27
Recently, a comprehensive diagnostic offering testing solution that combines the 23-GEP and 35-GEP has been introduced to provide additional clarity in characterizing ambiguous lesions.30 The combination test intends to take advantage of the strengths of both assays. As the 23-GEP offers well-studied, including validation, clinical utility and patient outcomes, all specimens initially undergo the 23-GEP test. The cases that receive intermediate result, which can be perceived as nonactionable, are further analyzed by 35-GEP, which has demonstrated low intermediate result rates.28,30,31 Goldberg et al31 compared the rates of actionable reports (benign or malignant test results) of the comprehensive diagnostic offering and 23-GEP testing alone. The study cohort included 738 archival biopsy samples of cutaneous melanocytic lesions consensus diagnosed by at least three independent dermatopathologists. All samples underwent the 23-GEP test, which reported nonactionable classification (intermediate result or technical failure) in 22.2 percent of the results. The samples with nonactionable results then were submitted for further testing with 35-GEP, which produced actionable results in additional 20.9 percent of the original cases. Thus, the comprehensive diagnostic offering workflow increased the rate of actionable results to 98.7 percent compared to 77.8 percent for the 23-GEP testing alone.31
DecisionDx-Melanoma (Castle Biosciences, Inc., Friendswood, Texas) is a GEP test that facilitates prognosis of melanoma as it uses RT-PCR methodology to quantify the expression of 31 genes from primary tumors. The 31-GEP test improves the accuracy in assessing the risk for positive sentinel lymph node (SLN) and recurrence. An algorithm is applied that provides results that classify Stage I-III melanoma patients into a low-risk (Class 1) or high-risk (Class 2) category of recurrence. The test divides patients into 4 classes: Class 1A (low risk of SLN positivity), Class 1B (low risk of melanoma recurrence), Class 2A (higher risk of SLN positivity) and Class 2B (high risk of melanoma recurrence).6 Several studies have demonstrated the capability of the 31-GEP test to improve risk assessment. Vetto et al32 conducted a study to determine if the test can be used to identify patients with low risk for SLN positivity (<5%) in the T1–T2 melanoma population who based on the American Joint Committee on Cancer (AJCC) guidelines would be considered for SLN biopsy. Two multicenter cohorts were used, a prospectively-tested cohort (n=1421) and a retrospective cohort (n=690). Results demonstrated that patients in age groups of 55–64 years and those aged 65 years or older with Class 1A profiles had SLN positivity rates of 4.9 percent (95% CI 2.3–9.2%) and 1.6 percent (95% CI 0.5–3.6%), respectively. Patients 55–64 years old and 65 years or older with Class 2B had SLN positivity rates of 30.8 percent (95% CI 9.1–61.4%) and 11.9 percent (95% CI 4.0–25.6%), respectively. Melanoma-specific survival was 99.3 percent for patients 55 years or older with Class 1A, T1–T2 tumors and 55.0 percent for Class 2B, SLN-positive, T1–T2 tumors.32 Another study conducted by Gastman et al. evaluated the risk prediction of the 31-GEP test within 3 low-risk populations of patients with cutaneous melanoma (CM): those who are SLN-negative, those with stage I to IIA tumors, and those with thin (≤ 1mm [T1]) tumors. The study included 690 patients pooled from three previous studies. Results demonstrated that the 31-GEP test identified 70 percent of SLN-negative patients who experienced metastasis as Class 2 and that the test is an independent predictor of recurrence.33 Various clinical utility studies have shown that the 31-GEP test strongly impacts clinical management decisions. A multicenter study conducted by Berger et al6 demonstrated clinical management changes in 82 out of 156 CM patients (53%). Similarly, another multicenter study conducted by Dillon et al. demonstrated that post-test management plans changed for 122 of 247 cases in the study when compared to pre-test plans (49%)34 Schuitevoerder et al35 conducted a study demonstrating that the 31-GEP test results accounted for 52 percewnt of the decision to manage clinically node negative cutaneous melanoma patients staged with SLN biopsy. Ferris et al. aimed to compare the accuracy of the 31-GEP test and AJCC Individualized Melanoma Patient Outcome Prediction Tool. One hundred and nine Stage I and 96 Stage II melanomas were included in the cohort to be evaluated. Results demonstrate that the 31-GEP test in combination with the AJCC test increase the sensitivity significantly, increasing the sensitivity to 88 percent for recurring melanomas and to 85 percent for distance metastasis.36 Furthermore, an intended decision study of 169 physicians conducted by Farberg et al. demonstrated 47–50% change in clinical management decisions for Class 2 results.37
Recently, there has been some controversy regarding the accuracy of the 31-GEP test performance studies. Greenhaw et al38 conducted a study to assess the accuracy of the prognostic value of the 31-GEP test. This study included a novel cohort of 211 (n = 1,479) cutaneous melanoma patients. Results demonstrated that the test identified patients with AJCC Stage I to III with high likelihood for recurrence and distant metastasis with a sensitivity of 76 percent (95% CI 71%-80%) for recurrence and 76 percent (95% CI 70%-82%) for distant metastasis.38 However, Marchetti et al39 argued that the meta-analysis of this study had a small sample size and methodological shortcomings, such as lack of a prespecified protocol, adjustment for confounders and missing data, meta-conflicts of interest and, incomplete risk of bias assessment (such as publication bias). In addition, a study conducted by Podlipnik et al40 evaluated the early prognostic accuracy of the 31-GEP test. The test was performed on 86 patients with AJCC stages IB and II to determine risk of recurrence. Results demonstrated that recurrence was identified in seven patients, all of which were in Class 2 by the 31-GEP test, representing 21.2 percent of the group. As a result, the 31-GEP test was termed as an accurate test to identify patients at early AJCC stages who are at greater risk of relapse. However, a study conducted by Marchetti et al39 demonstrated opposite findings. The study aimed to systematically assess the performance of the test in patients with AJCC Stage I or stage II disease. Five studies assessing the 31-GEP test results in a total of 835 patients were evaluated. Results demonstrated that the test correctly classified recurrence in 29 percent of patients with Stage I disease and 82 percent of patients with Stage II disease; however, the test results varied by AJCC stage and were unreliable in predicting recurrence for patients with AJCC Stage I disease.41
Conclusion
Over the last few decades, the incidence of melanoma in the United States has been rising. However, the diagnosis of melanoma with unaided visual inspection remains a challenge. New diagnostic and prognostic technologies demonstrate promising results for early and accurate detection of melanocytic lesions. EIS and PLA technologies may facilitate biopsy decisions for physicians. RCM, adjunct with other technologies, has the potential to improve diagnostic accuracy for melanoma. The 23-GEP test may aid in the distinction between melanocytic lesions and benign nevi. The 35-GEP test may potentially refine diagnoses of challenging melanocytic lesions. The new comprehensive diagnostic offering testing that combines the 23-GEP and 35-GEP may provide additional clarity in ambiguous cases. The 31-GEP test intends to improve the prognosis for melanoma. However, each of the tools has its limitations; thus, it is important for clinicians to review the latest studies on the technologies to ensure the appropriate diagnosis, management, and prognosis of melanoma.
References
- American Cancer Society. Facts & Figures 2021. American Cancer Society. Atlanta, Ga. 2021.
- Ollmar S, Grant S. Nevisense: improving the accuracy of diagnosing melanoma. Melanoma Manag. 2016 Jun;3(2):93–96.
- Ferris LK, Moy RL, Gerami P, et al. Noninvasive Analysis of High-Risk Driver Mutations and Gene Expression Profiles in Primary Cutaneous Melanoma. J Invest Dermatol. 2019 May;139(5):1127–1134.
- González S, Gilaberte‐Calzada Y. In vivo reflectance-mode confocal microscopy in clinical dermatology and cosmetology. Int J Cosmet Sci. 2008;30(1):11–17.
- Clarke LE, Warf MB, Flake DD, et al. Clinical validation of a gene expression signature that differentiates benign nevi from malignant melanoma. J Cutan Pathol. 2015;42(4):244–252.
- Berger AC, Davidson RS, Poitras JK, et al. Clinical impact of a 31-gene expression profile test for cutaneous melanoma in 156 prospectively and consecutively tested patients. Curr Med Res Opin. 2016 Sep 1;32(9):1599–1604.
- Scibase. Neviscence Clinical Reference Guide. Published October 4, 2014. Accessed_August 24, 2021. https://scibase.com/wp-content/uploads/2017/11/Clinical-Reference-_Guide-1.pdf.
- Malvehy J, Hauschild A, Curiel-Lewandrowski C, et al. Clinical performance of the Nevisense system in cutaneous melanoma detection: an international, multicentre, prospective and blinded clinical trial on efficacy and safety. Br J Dermatol. 2014 Nov;171(5):1099–1107.
- Svoboda RM, Prado G, Mirsky RS, Rigel DS. Assessment of clinician accuracy for diagnosing melanoma on the basis of electrical impedance spectroscopy score plus morphology versus lesion morphology alone. J Am Acad Dermatol. 2019 Jan 1;80(1):285–287.
- Rocha L, Menzies SW, Lo S, et al. Analysis of an electrical impedance spectroscopy system in short-term digital dermoscopy imaging of melanocytic lesions. Br J Dermatol. 2017;177(5):1432–1438.
- Brouha B, Ferris L, Skelsey M, et al. Real-World Utility of a Non-Invasive Gene Expression Test to Rule Out Primary Cutaneous Melanoma: A Large US Registry Study. J Drugs Dermatol. 2020 Mar 1;19(3):257–262.
- Shah A, Hyngstrom J, Florell SR, et al. Use of the Pigmented Lesion Assay to rapidly screen a patient with numerous clinically atypical pigmented lesions. JAAD Case Rep. 2019 Dec 1;5(12):1048–1050.
- Gerami P, Yao Z, Polsky D, et al. Development and validation of a noninvasive 2-gene molecular assay for cutaneous melanoma. J Am Acad Dermatol. 2017 Jan 1;76(1):114–120.e2.
- Ferris LK, Gerami P, Skelsey MK, et al. Real-world performance and utility of a noninvasive gene expression assay to evaluate melanoma risk in pigmented lesions. Melanoma Res. 2018 Oct;28(5):478–482.
- Ferris LK, Rigel DS, Siegel DM, et al. Impact on clinical practice of a non-invasive gene expression melanoma rule-out test: 12-month follow-up of negative test results and utility data from a large US registry study. Dermatol Online J. 25(5):8.
- Ferris LK, Jansen B, Ho J, et al. Utility of a Noninvasive 2-Gene Molecular Assay for Cutaneous Melanoma and Effect on the Decision to Biopsy. JAMA Dermatol. 2017 Jul 1;153(7):675.
- DermTech Introduces PLAplus with Improved Sensitivity for Early Detection of Melanoma. 2021. Available at: https://investors.dermtech.com/node/8086/pdf. Accessed August 24, 2021.
- Jackson Cullison SR, Jansen B, Yao Z, et al. Risk stratification of severely dysplastic nevi by non-invasively obtained gene expression and mutation analyses. SKIN J Cutan Med. 2020 Mar 8;4(2):124.
- Cullison S, Ferris L, Yao Z, et al. Combining DNA and RNA Analyses Enhances Non-Invasive Early Detection of Cutaneous Melanoma. SKIN J Cutan Med. 2020 Oct 27;4(6):s126–s126.
- Waddell A, Star P, Guitera P. Advances in the use of reflectance confocal microscopy in melanoma. Melanoma Manag. 2018;5(1):MMT04
- Lan J, Wen J, Cao S, et al. The diagnostic accuracy of dermoscopy and reflectance confocal microscopy for amelanotic/hypomelanotic melanoma: a systematic review and meta-analysis. Br J Dermatol. 2020;183(2):210–219.
- Xiong YQ, Ma SJ, Mo Y, et al. Comparison of dermoscopy and reflectance confocal microscopy for the diagnosis of malignant skin tumours: a meta-analysis. J Cancer Res Clin Oncol. 2017 Sep 1;143(9):1627–1635.
- Pezzini C, Kaleci S, Chester J, et al. Reflectance confocal microscopy diagnostic accuracy for malignant melanoma in different clinical settings: systematic review and meta-analysis. J Eur Acad Dermatol Venereol. 2020;34(10):2268–2279.
- Clarke LE, Flake DD, Busam K, et al. An independent validation of a gene expression signature to differentiate malignant melanoma from benign melanocytic nevi. Cancer. 2017;123(4):617–628.
- Ko JS, Matharoo-Ball B, Billings SD, et al. Diagnostic Distinction of Malignant Melanoma and Benign Nevi by a Gene Expression Signature and Correlation to Clinical Outcomes. Cancer Epidemiol Prev Biomark. 2017 Jul 1;26(7):1107–1113.
- Ko JS, Clarke LE, Minca EC, et al. Correlation of melanoma gene expression score with clinical outcomes on a series of melanocytic lesions. Hum Pathol. 2019 Apr 1;86:213–221.
- Estrada S, Shackelton J, Cleaver N, et al. Development and Validation of a Diagnostic 35-Gene Expression Profile Test for Ambiguous or Difficult-to-Diagnose Suspicious Pigmented Skin Lesions. SKIN J Cutan Med. 2020 Oct 27;4(6):506–522.
- Cockerell C, Goldberg M, Estrada S, et al. Performance of a 35-Gene Expression Profile Test in Suspicious Pigmented Lesions of the Head and Neck. SKIN J Cutan Med. 2021;5(1):s1–s1.
- Farberg A, Ahmed K, Bailey C, et al. A 35-Gene Expression Profile Test for use in Suspicious Pigmented Lesions Impacts Clinical Management Decisions of Dermatopathologists and Dermatologists. SKIN J Cutan Med. 2020 Oct 27;4(6):523–533.
- Castle Biosciences. MyPath Melanoma and DiffDx-Melanoma Overview. Castle Biosciences, Inc. https://castlebiosciences.com/products/mypath-melanoma-and-diffdx-melanoma/. Published February 27, 2022. Accessed May 13, 2022.
- Goldberg M, Siegel J, Russell B, et al. A comprehensive diagnostic offering workflow increases the rate of actionable results of the 23- and 35-gene expression profile tests for use as ancillary diagnostic tools for difficult-to-diagnose melanocytic lesions. SKIN J Cutan Med. 2021 Nov 5;5(6):s79–s79.
- Vetto JT, Hsueh EC, Gastman BR, et al. Guidance of sentinel lymph node biopsy decisions in patients with T1–T2 melanoma using gene expression profiling. Future Oncol. 2019 Jan 29;15(11):1207–1217.
- Gastman BR, Gerami P, Kurley SJ, et al. Identification of patients at risk of metastasis using a prognostic 31-gene expression profile in subpopulations of melanoma patients with favorable outcomes by standard criteria. J Am Acad Dermatol. 2019 Jan 1;80(1):149–157.e4.
- Dillon LD, Gadzia JE, Davidson RS, et al. Prospective, Multicenter Clinical Impact Evaluation of a 31-Gene Expression Profile Test for Management of Melanoma Patients. SKIN J Cutan Med. 2018 Mar 9;2(2):111–121.
- Schuitevoerder D, Heath M, Cook RW, et al. Impact of Gene Expression Profiling on Decision-Making in Clinically Node Negative Melanoma Patients after Surgical Staging. J Drugs Dermatol JDD. 2018 Feb 1;17(2):196–199.
- Ferris LK, Farberg AS, Middlebrook B, et al. Identification of high-risk cutaneous melanoma tumors is improved when combining the online American Joint Committee on Cancer Individualized Melanoma Patient Outcome Prediction Tool with a 31-gene expression profile–based classification. J Am Acad Dermatol. 2017 May 1;76(5):818–825.e3.
- Farberg AS, Glazer AM, Winkelmann RR, et al. Assessing Genetic Expression Profiles in Melanoma Prognosis. Dermatol Clin. 2017 Oct 1;35(4):545–550.
- Greenhaw BN, Covington KR, Kurley SJ, et al. Molecular risk prediction in cutaneous melanoma: A meta-analysis of the 31-gene expression profile prognostic test in 1,479 patients. J Am Acad Dermatol. 2020 Sep;83(3):745–753.
- Marchetti MA, Dusza SW, Bartlett EK. Problematic methodology in a systematic review and meta-analysis of DecisionDx-Melanoma. J Am Acad Dermatol. 2020 Nov 1;83(5):e357–358.
- Podlipnik S, Carrera C, Boada A, et al. Early outcome of a 31-gene expression profile test in 86 AJCC stage IB-II melanoma patients. A prospective multicentre cohort study. J Eur Acad Dermatol Venereol. 2019;33(5):857–862.
- Marchetti MA, Coit DG, Dusza SW, et al. Performance of Gene Expression Profile Tests for Prognosis in Patients With Localized Cutaneous Melanoma: A Systematic Review and Meta-analysis. JAMA Dermatol. 2020 Sep 1;156(9):953–962.