J Clin Aesthet Dermatol. 2023;16(2):55–59.
J Clin Aesthet Dermatol. 2023;16(2):55–59.
by Abd El-Aziz El-Taweel, MD; Amira Abdelrahman, MD; Hebatallah Hegazy, MSc; and Rehab Salem, MD
Dr. El-Taweel is with Faculty of Medicine at 6th of October University in Cairo, Egypt. Drs. Abdelrahman, and Salem are with the Faculty of Medicine at Benha University in Banha, Egypt. Dr. Hegazy is with Ministry of Health Hospital in Alexandria, Egypt.
ABSTRACT: Background. Granzyme B (GZMB) gene is related to human immunity and is considered as one of the genes indulged in vitiligo.
Objective. To evaluate the association between GZMB (R55Q) and (P94Q) gene polymorphisms with vitiligo development in a sample of vitiligo Egyptian patients.
Methods. This study was a case-control study which included 100 non-segmental vitiligo patients as well as a control group consisted of 100 healthy, sex and age matched vitiligo free individuals. The polymorphism of GZMB gene at (R55Q) and (P94A) were analyzed by polymerase chain reaction.
Results. P94Q and R55Q gene polymorphisms were significantly associated with higher VASI scores.
Conclusion. GZMB (Q55R) and P94A) gene polymorphisms are associated with the susceptibility to develop vitiligo in the Egyptian population and could help predicting more extensive forms of the disease.
Vitiligo is a chronic amelanocytic skin disorder which is usually acquired. The main pathology in the vitiliginous patches is the complete melanocytes loss, however, the exact cause of this loss is still under debate. Depigmentation may affect both skin and hair to different extents.1,2
Several theories to explain the occurrence of this disorder were proposed. They include genetic predisposition, autoimmunity, autocytotoxicity, neurogenic elements, microenviromental factors, viral infection, melanocytes apoptosis and adhesion disorders.3 The most accepted theory of vitiligo pathogenesis is the occurrence of one or more of the mentioned hypotheses in a genetically predisposed person.4
Many genetic studies are directed to map the susceptibility loci for vitiligo in order to understand the genetic background of the diseases more clearly. However, these studies could not provide the full explanation of the disorder5-9, so this field needs more attention and more studies are required.
Granzyme B enzyme; a serine protease, is secreted with perforin by natural killer cells and cytotoxic lymphocytes. They act together to induce apoptosis of cells.10 Perforin helps the internalization of Granzyme B in the target cell, then Granzyme B induces apoptosis of this cell by caspase- dependent and or -independent pathways.11,12 GZMB can cleave melanocytic proteins attributing to the initiation and propagation of autoimmune reaction towards self melanocytes.13
GranzymeB (GZMB) gene is located at 14q12 and it has five exons. It is related to the immune system functions, so it was proposed to be one of the genes involved in the etiopathogenesis of vitiligo.14,15
Xu et al16 studied the relation between 15 GZMB SNPs with the development of vitiligo and the progress in its course in a sample of Chinese Han ancestry. They concluded that this gene polymorphism may play a role in vitiligo cases in that race.
This work was designed to evaluate the association between GZMB (R55Q) and (P94Q) gene polymorphisms with vitiligo occurrence and to assess the clinical significance of these polymorphisms among a sample of Egyptian vitiligo patients.
Methods
This case control study included 100 patients with non-segmental vitiligo as well as a control group consisting of 100 healthy, sex, and age-matched vitiligo individuals without vitiligo. Patients were recruited from the outpatient clinic and the phototherapy unit of Dermatology, Venereology, and Andrology Department of Benha University Hospitals. An informed consent was obtained from all participants after explanation of the study and before collecting blood samples. The protocol of this study obtained the approval of the local ethics committee on research involving human subjects of Benha Faculty of Medicine (Ethical Approval: Ms.27.1.2019).
Subjects with any of the following criteria were excluded from the study; other autoimmune or inflammatory skin or systemic disease, chronic infectious diseases, malignancy and chronic systemic diseases (e.g. hepatic, renal, cardiac, etc).
All patients in our sample were subjected to complete history taking, complete cutaneous examination to evaluate the extent and clinical type of vitiligo, the presence of halo nevi or leukotrichia and the involvement of mucous membranes, eye and ears. Vitiligo severity was assessed by Vitiligo area severity index (VASI).17
Genetic analyses. Samples of peripheral venous blood (2mL) were collected under complete aseptic conditions in EDTA containing tubes from all participants. DNA was isolated from all samples using G-Spin DNA Extraction kit (Cat. No.17045) according to the manufacturer’s instructions and stored at -20°c.
The polymorphism of GZMB gene at (R55Q) and (P94A) were analyzed by polymerase chain reaction followed by DNA purification. The DNA was amplified by PCR using appropriate forward primer: 5’- – AGT GTT TCC AGG AGG GTG TG– 3’ and reverse primer: 5′ –TTT TCC TTC AGG GGA GAT CA-3′ for GZMB (R55Q) and Forward primer: 5′ –CCT CAC CTG CAG TAG CAT GA-3′ and reverse primer: 5′ –CCC ATC CTC ACT CTG CTC TC-3′ for GZMB (P94A).
The final volume of 25μL of a mixture containing 10 nanogram of DNA mixed with 2.5μL of gene-specific primers and probes and 12.5μL of PCR universal master mix. Thermal cycle parameters were as follows: eight minutes denaturing at 95°C, followed by 40 cycles involving denaturation at 95°C for 1.5 min, and annealing/extension at 60°C for one min and extension at 72°C for 1.5 min. Sample duplication, negative (-ve) and positive (+ve) control of a sequenced sample were used to confirm the accuracy of genotyping.
Purified DNA sequencing was performed using ABI Prism 3.1 automated sequencer (Applied Biosystems, Foster City, CA, USA) and chromaspro software 1.34 was used to analyze data.
Data management and analysis. Data were displayed in tables and statistically analyzed by the Statistical package for Social Science (IBM Corp. Released 2017.IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY:IBM Corp.). Tests were chosen according to the type of data obtained for each parameter. The normality of data distribution was tested by Shapiro-Wilk test. Significant data was considered to be nonparametric.
Deviations from Hardy–Weinberg (HW) equilibrium expectations were determined using the chi-squared test. Haplotypes were estimated by the version 4.2 of the HaploView program. All reported p values were two-tailed and if p value is less than 0.05, the result was considered to be significant.
Results
There was an insignificant difference between control subjects and vitiligo patients regarding age (43.6±8.4 versus 40.2±11.3, p=0.104 ) and sex (36 male control subjects “36%” versus 34 male patients “34%”,p = 0.834).
The mean age of vitiligo onset was 19.6±6.1 years and the mean disease duration was 20.4±5.3 years. Twenty patients (20%) had positive family history of vitiligo. Among the studied cases, 36 patients had focal vitiligo (36%), 72 patients had acrofacial vitiligo (72%), 24 patients had vitiligo vulgaris (24%) and four patients had universal vitiligo (4%). Hair was affected in 12 patients (12%). The mean VASI score was 47.3±14.2.
Genetic studies. Both SNPs are located within granzyme B gene at long arm of 14 chromosome, with less than one kilobase apart (571 bases). GZMB (P94Q) (rs11539752) had C and G alleles.G Allele change from GCC to CCC results in amino acid change from proline into alanine. G allele was considered as the wild allele. GZMB (R55Q) (rs8192917) had A and G alleles. An Allele change from CAA to CGA results in amino acid change from arginine into glycine. A allele was considered as the wild allele.18
This sample of individuals was selected randomly. Applying Hardy Weinberg (HW) equation, revealed that P94Q and R55Q genotypes in control subjects as well as in cases groups were in HW equilibrium (Table 1).
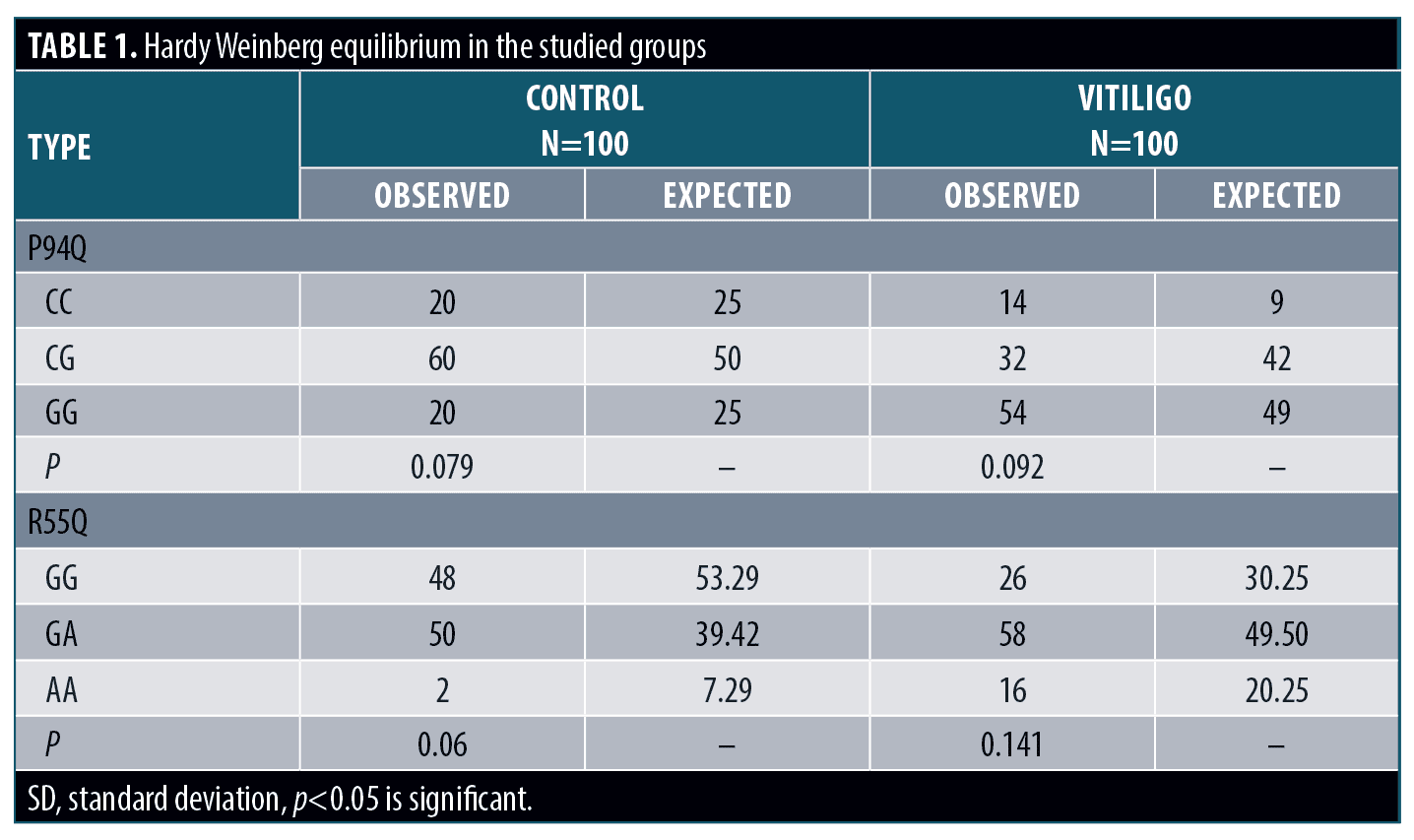
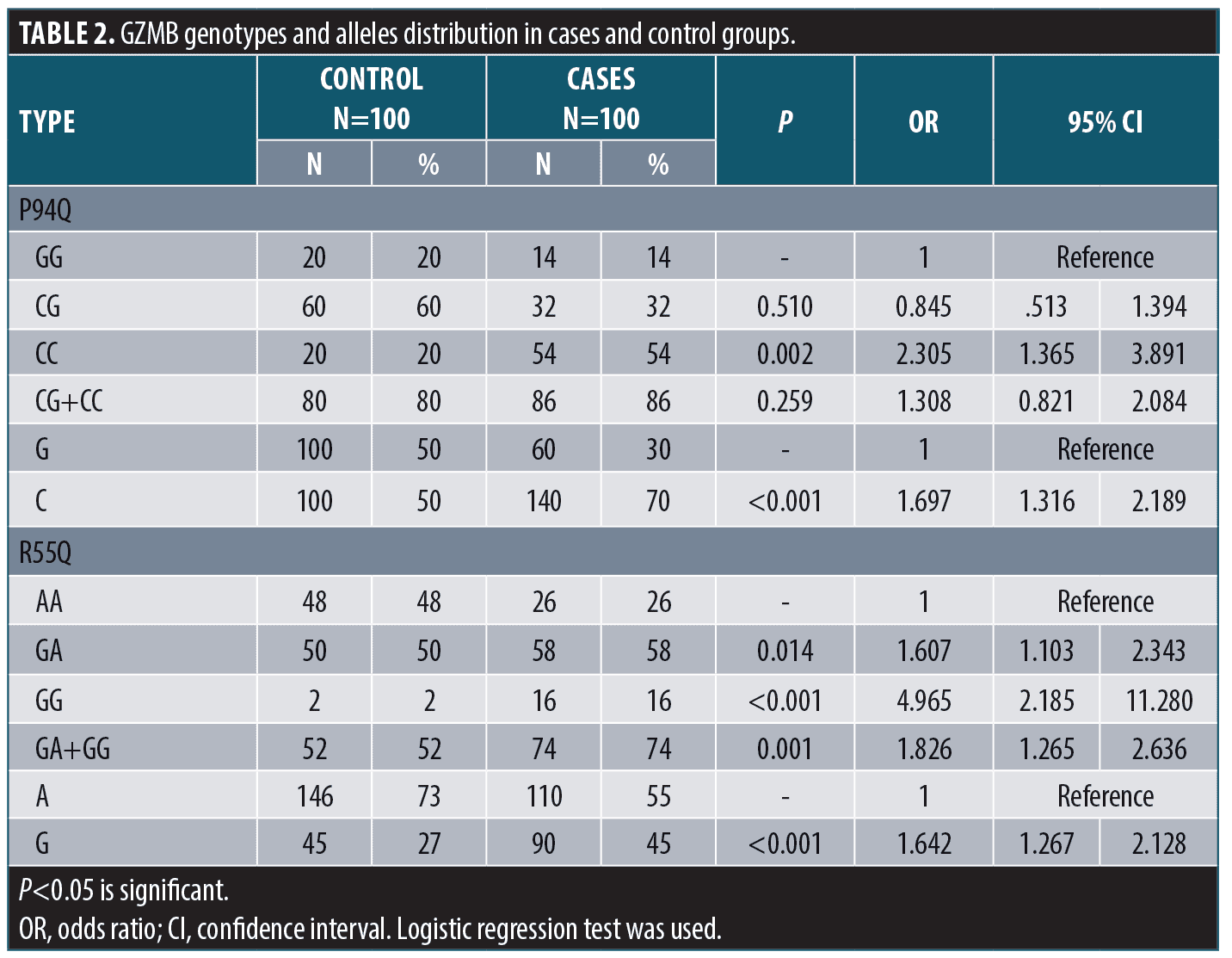
Genotypes and alleles distribution. Regarding P94Q; CC genotype and C allele showed significantly higher frequency in cases when compared to the control group (p<0.05 for each); with increased risk to develop vitiligo (OR>1for each). Regarding R55Q; GG genotype and G allele showed significantly higher frequency in cases when compared to the control group (p<0.05 for each); with increased risk to develop vitiligo (OR>1 for each) (Table 2).
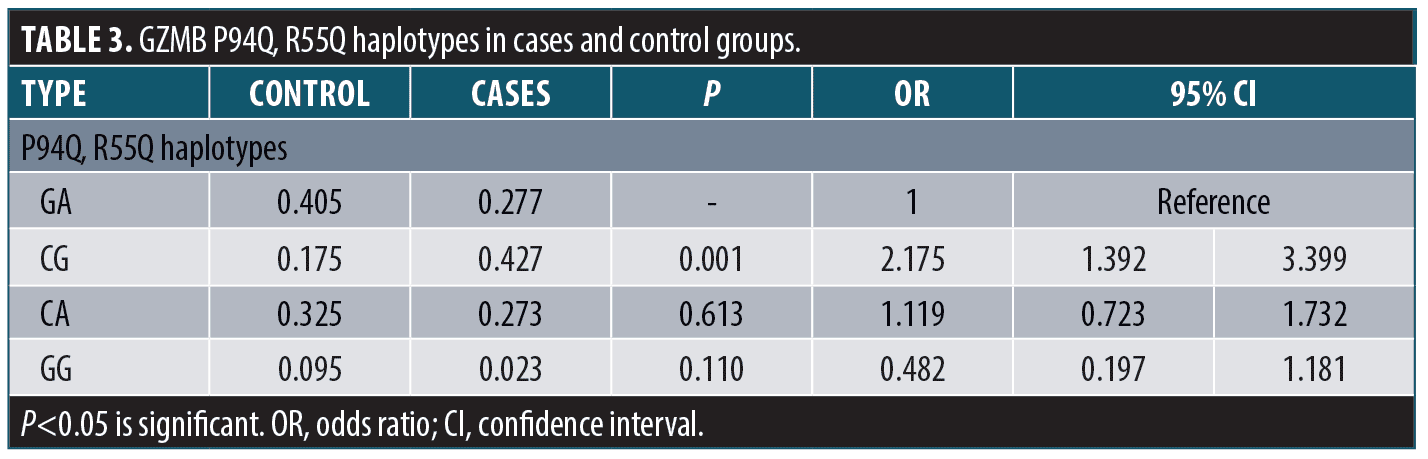
The granzyme B P94Q, R55Q haplotypes were calculated; CG haplotype (combined polymorphism at both SNPS) was significantly associated with risk of vitiligo occurrence (p=0.001, OR=2.175) (Table 3).
Linkage disequilibrium refers to the non-random association of alleles at ≥ 2 loci in a general population. D’ varies between zero (no disequilibrium) and one (maximum disequilibrium). In studied control group, D’=0.61; in studied cases, D’=0.90.
P94Q polymorphism and the study variables. There was insignificant difference in P94Q genotypes distribution between patients and control groups regarding demographic, history or clinical data except for VASI score. P94Q gene polymorphism was associated with significantly higher VASI scores (Table 4).
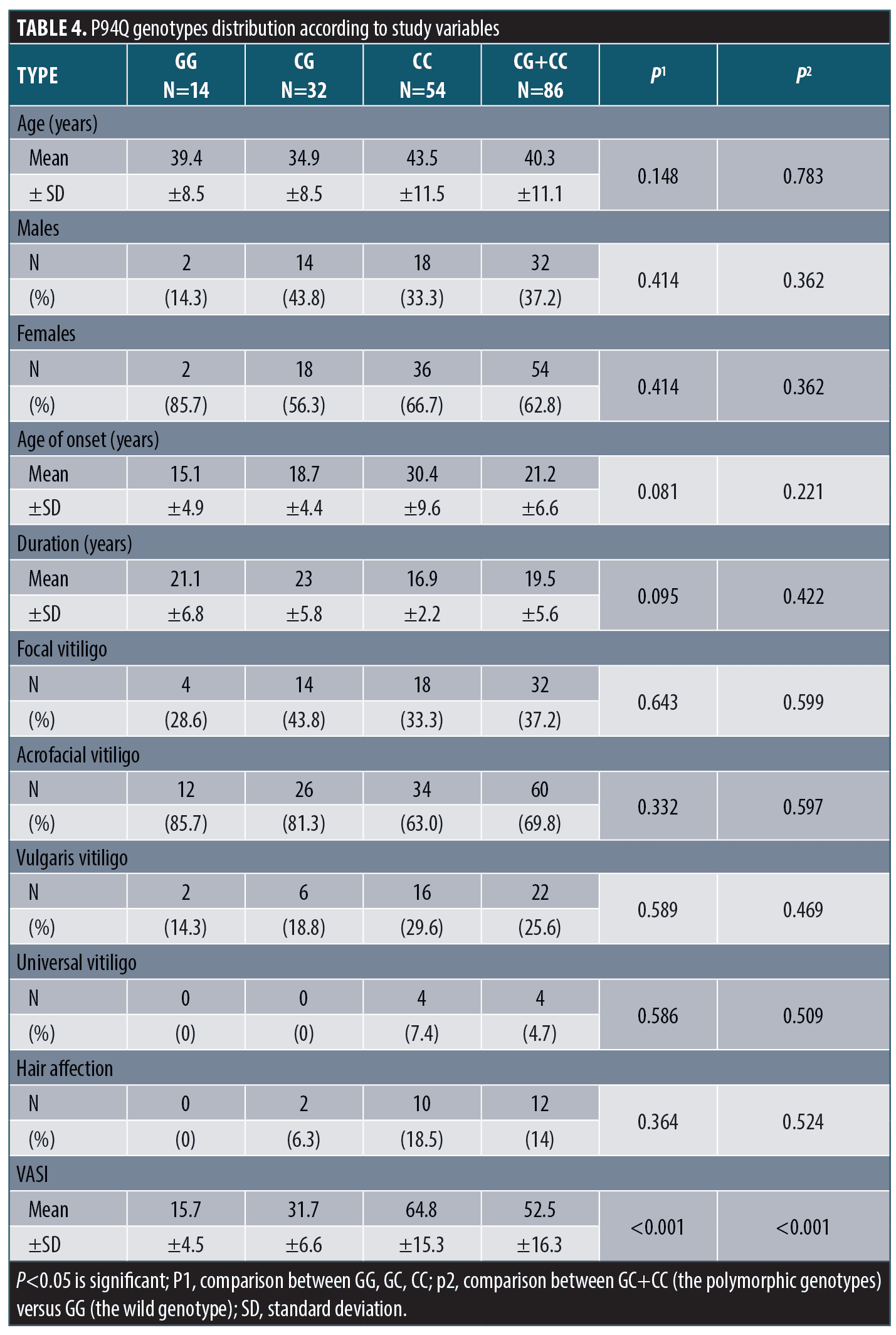
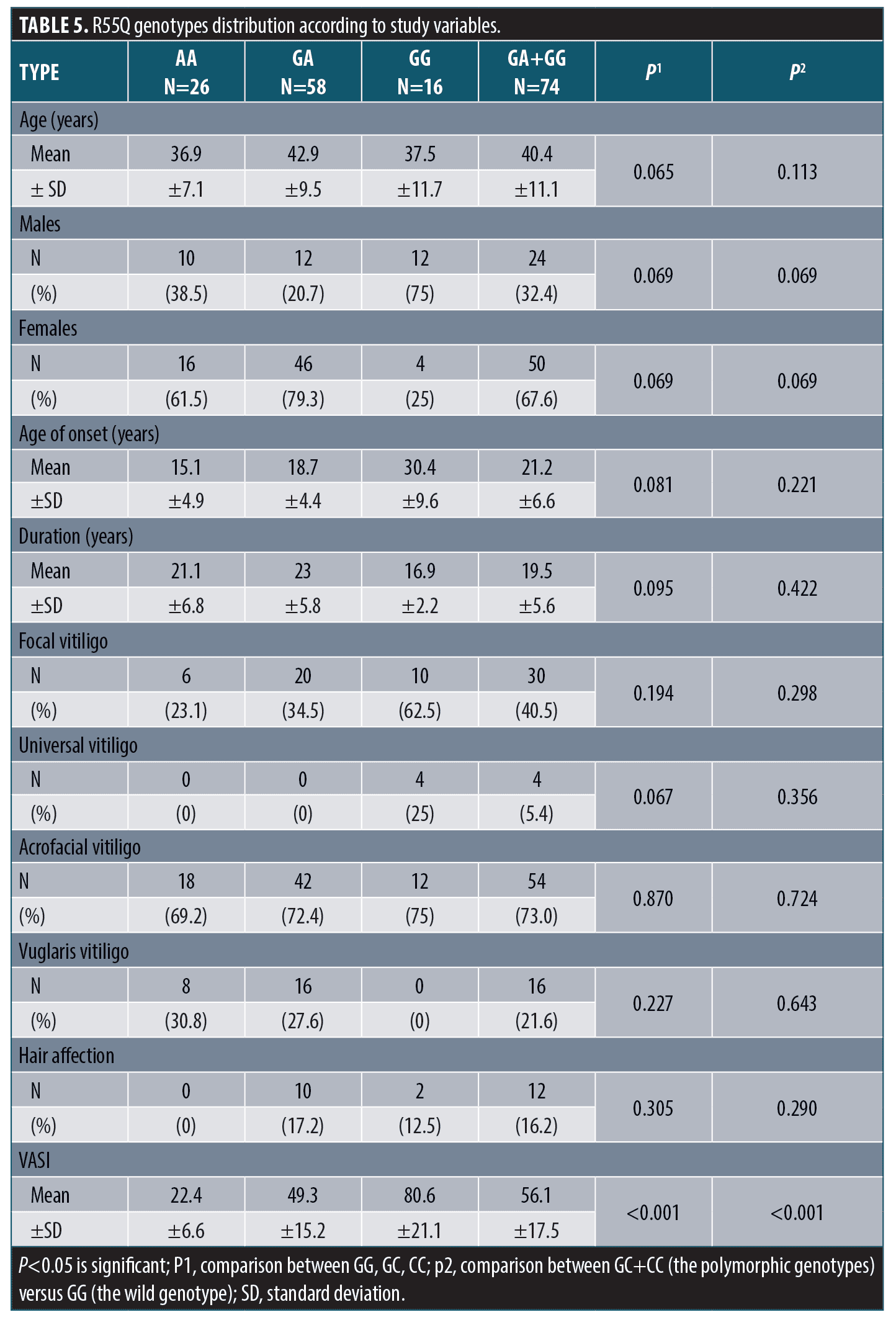
R55Q polymorphism and the study variables. There was insignificant difference in R55Q genotypes distribution between patients and control groups regarding demographic, history or clinical data except for VASI score. R55Q gene polymorphism was associated with significantly higher VASI score (Table 5).
Discussion
GzmB (P94Q) had C and G alleles, G is the wild allele. Allele change from CCC to GCC results in missense amino acid change from proline into alanine. GzmB (R55Q) had A and G alleles, A is the wild allele. Allele change from CGA to CAA results in missense amino acid change from arginine into glycine. Both SNPs are located within granzyme B gene at long arm of chromosome14, with less than one kilobase apart (571 bases). Applying Hardy Weinberg equation, revealed that P94Q and R55Q genotypes in control as well as in cases groups in this study were in HW equilibrium.19
In the current work, P94Q CC genotype and C allele showed significantly higher frequency in cases when compared to control groups with an increased risk to develop vitiligo. Regarding R55Q; GG genotype and G allele showed significantly higher frequency in cases when compared to control group with an increased risk to develop vitiligo too. The combined polymorphisms at both SNPs is associated with more increased risk of vitiligo development. The present results agree with Jeong et al20 and Xu et al16 who reported the association of the same SNPs with the increased risk of vitiligo development in a Korean and Chinese vitiligo patients samples, respectively.
To the best of our knowledge, this is the first time to study the impact of these SNPs on the clinical aspect of non-segmental vitiligo. The current work highlighted the association of these polymorphisms with significantly higher vitiligo severity (higher VASI scores). These results suggest an important role of GNMB gene polymorphisms in vitiligo development and in increasing its severity.
The impact of GZMB (Q55R) polymorphism on the levels and activity of the enzymes is a matter of debate. This SNP may increase granzyme B expression21, or affect its cytolytic activity.22 However, Zaitsu et al23 and Sun et al24 reported that this GZMB gene polymorphism does not affect the enzyme function. GZMB P94A gene polymorphism may also be associated with changes in the enzyme activity with subsequent defects in the immune systems responses.25
How granzyme B enzyme would become involved in the pathogenesis of vitiligo is not clear yet, however, it seems that this cytolytic enzyme is somehow involved in more than one of the hypotheses proposed to explain vitiligo development. Granzyme B has cleavage sites on many melanocytes autoantigens that are targeted in vitiligo e.g. TYR , TYRP1, TYRP2, SOX10, and PMEL.26 Moreover, the blood and tissue levels of granzyme B producing cells were significantly increased in NSV patients.27 Granzyme B may also be involved in generating oxidative stress during apoptosis,28 impairing neuronal differentiation29 and inducing melanocytes defects or self destruction30 which are all considered as an etiopathogenic mechanism of vitiligo initiation or propagation.
Conclusion
From the results of present study, it is concluded that GZMB (Q55R) and P94A) gene polymorphisms might be associated with the susceptibility to develop vitiligo in the Egyptian population and could help predicting more extensive forms of the disease.
References
- Boniface K, Jacquemin C, Darrigade AS et al. Vitiligo skin is imprinted with resident memory CD8 T cells expressing CXCR3. J Invest Dermatol. 2018 Feb;138(2):355–364
- Bergqvist C, Ezzedine K. Vitiligo: A Review. Dermatology. 2020; 236(6):571–592.
- Taïeb A, Picardo M. Clinical practice. Vitiligo. N Engl J Med. 2009 Jan 8;360(2):160–169.
- Spritz RA. Recent progress in the genetics of generalized vitiligo. J Genet Genomics. 2011;38(7):271–278.
- Spritz RA, Andersen GH. Genetics of Vitiligo. Dermatol Clin. 2017; 35(2):245–255.
- Monib KME, Sabry HH, Hussein MS, et al. Factors affecting vitiligo response to treatment: do MiRNA 196a2C/T gene polymorphism and serum tyrosinase levels have any role? J Dermatolog Treat. 2020 Aug 24:1–5.
- Salem RM, Abdelrahman AMN, Abd El-Kareem HM, et al. DEFB1 gene polymorphisms modify vitiligo extent and response to NB-UVB phototherapy. Dermatol Ther. 2021 May;34(3):e14921.
- Sorour NE, Abd El-Kareem HM, Ibrahim AE, et al. Nuclear Factor Erythroid-2-related Factor 2 Gene Polymorphisms in Vitiligo. J Clin Aesthet Dermatol. 2021 Jun;14(6):14–17.
- El-Esawy FM, Abd El-Kareem HM, Mohamady AA, et al. Proteasome Subunit Beta Type-8 (PSMB8) Gene Polymorphisms in Vitiligo: A Possible Predictor of Auditory Involvement. J Clin Aesthet Dermatol. 2021 Dec;14(12):30–35.
- Turner CT, Zeglinski MR, Richardson KC, et al. Granzyme K Expressed by Classically Activated Macrophages Contributes to Inflammation and Impaired Remodeling. J Invest Dermatol. 2019;139(4):930–939.
- Thiery and Lieberman et al. Perforin: a key pore-forming protein for immune control of viruses and cancer. Subcell Biochem. 2014;80:197–220.
- Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol. 2015; 15(6):388–400.
- Spritz RA. Modern vitiligo genetics sheds new light on an ancient disease. J Dermatol. 2013; 40(5):310–318.
- Ezzedine K, Eleftheriadou V, Whitton M, et al. Lancet. 2015; 386(9988):74–84.
- Turner Lim & Granville et al. Granzyme B in skin inflammation and disease. Matrix Biol. 2019 Jan;75-76:126–140.
- Xu M, Liu Y, Liu Y, et al. Genetic polymorphisms of GZMB and vitiligo: A genetic association study based on Chinese Han population. Sci Rep. 2018;8(1):13001.
- Hamzavi I, Jain H, McLean D, et al. Parametric modeling of narrowband UV-B phototherapy for vitiligo using a novel quantitative tool: the Vitiligo Area Scoring Index. Arch Dermatol. 2004 Jun;140(6):677–683.
- National Center for Biotechnology Information database, 2019. Retrieved from https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?do_not_redirect&rs=rs8192917
- National Center for Biotechnology Information (NCBI). https://www.ncbi.nlm.nih.gov/gene/3002#gene-expression. Last updated on 18 July, 2022.
- Jeong KH, Kim SK, Seo JK, et al. Association of GZMB polymorphisms and susceptibility to non-segmental vitiligo in a Korean population. Sci Rep. 2021;11(1):397.
- Girnita DM, Webber SA, Brooks MM, et al. Genotypic variation and phenotypic characterization of granzyme B gene polymorphisms. Transplantation. 2009;87:1801–1806.
- McIlroy D, Cartron PF, Tuffery P, et al. A triple-mutated allele of granzyme B incapable of inducing apoptosis. Proc Natl Acad Sci USA. 2003;100:2562–7.
- Zaitsu M, Yamamoto K, Ishii E, et al. High frequency of QPY allele and linkage disequilibrium of granzyme-B in Epstein-Barr-virus-associated hemophagocytic lymphohistiocytosis. Tissue Antigens. 2004;64:611–615.
- Sun J, Bird CH, Thia KY, et al. Granzyme B encoded by the commonly occurring human RAH allele retains pro-apoptotic activity. J Biol Chem. 2004;279:16907-16911.
- Gaafar A, Aljurf MD, Al-Sulaiman A, et al. Defective gammadelta T-cell function and granzyme B gene polymorphism in a cohort of newly diagnosed breast cancer patients. Exp Hematol. 2009;37:838–848.
- Darrah E, Rosen A. Granzyme B cleavage of autoantigens in autoimmunity. Cell Death Differ. 2010 Apr;17(4):624–632.
- Dwivedi M, Laddha NC, Arora P, et al. Decreased regulatory T-cells and CD4(+) /CD8(+) ratio correlate with disease onset and progression in patients with generalized vitiligo. Pigment Cell Melanoma Res. 2013 Jul;26(4):586–591.
- Yang L, Wei Y, Sun Y, et al. Interferon-gamma Inhibits Melanogenesis and Induces Apoptosis in Melanocytes: A Pivotal Role of CD8+ Cytotoxic T Lymphocytes in Vitiligo. Acta Derm Venereol. 2015 Jul;95(6):664–670.
- Wang CQ, Cruz-Inigo AE, Fuentes-Duculan J, et al. Th17 cells and activated dendritic cells are increased in vitiligo lesions. PLoS One. 2011;6(4):e18907.
- Wang Y, Li S, Li C. Perspectives of New Advances in the Pathogenesis of Vitiligo: From Oxidative Stress to Autoimmunity. Med Sci Monit. 2019 Feb 6;25:1017–1023.